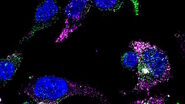
Consideration for optimal fluorescence live-cell imaging
Keeping your sample under physiological conditions:
The most important thing to take into account for your experiment is keeping your sample alive and healthy during sample preparation and imaging. Thus, it is critical to maintain the sample in an environment that resembles as closely as possible physiological conditions, i.e., temperature, pH, oxygen levels, and other important factors.
Minimum stress from light exposure:
Exposing live cells to light causes phototoxicity and changes in the physiology of your specimen, so that you might get results that do not resemble what actually happens in a healthy organism. To avoid phototoxicity as much as possible, widefield microscopes are often the technology of choice for imaging live specimens. They illuminate the sample with low doses of LED light to excite fluorophores and image acquisition is faster than other microscopy methods, such as confocal imaging.
Labeling of your sample:
When imaging live specimens, fluorescent proteins are normally the tool of choice to tag the specimen structures where the events you want to study happen, as these proteins are usually less toxic compared to other labels and circumvent the issue of antibodies not being able to enter the cells. Fluorescent protein technology relies on a genetically encoded fluorophore that can be used for the tracking of tagged biomolecules in living cells, their interactions, etc. Using multiple fluorescent proteins simultaneously, known as multicolor or multiplex imaging, allows you to observe multiple cell structures and processes in synchrony, providing more physiologically relevant results and, thus, adding context to measurements (e.g., live dead assays with additional markers or doing multicolor vesicle observation experiments). To learn more about how to perform multicolor imaging in live samples, read this article: Considerations for Multiplex Live Cell Imaging.
Methods employed for live-cell imaging
There is a wide range of microscopy techniques applied for live-cell imaging. Often, the growth of cells, cell aggregates, or cell movement is observed over time using compound microscopes and methods like phase contrast and differential interference contrast (DIC). Time- lapse imaging of larger specimens, such as developing zebrafish embryos, is usually performed with stereo microscopes or macroscopes.
This article focuses on fluorescence microscopy techniques that have become more prominent in the last few decades. This short overview covers commonly used quantitative methods, as well as photomanipulation techniques, that are commonly used with widefield microscopes.
Ion imaging – observing changes in ion concentrations
As the ion composition in the cytosol of cells determines many crucial functions, such as excitability of neurons, gene transcription, and cell movement, just to name a few, the regulation of intracellular ions in both spatial and temporal terms is of major interest for life science research. Ion imaging (calcium, chloride, magnesium) is a common method that uses either fluorescent dyes or proteins especially designed to change their emission behavior upon calcium binding. This allows researchers to observe dynamic changes in the ion concentrations of cells. Also, the imaging of intracellular pH-levels or voltage is possible with special fluorescent dyes. A special technique for detecting changes in ion levels, pH levels, or voltage is ratiometric imaging. This refers to a group of methods that allow an exact determination of changes in cells, e.g., the intracellular calcium concentration, instead of monitoring relative changes as it is done in non-ratiometric methods.
TIRF – observing processes close to the cell membrane
Total internal reflection fluorescence or TIRF is a special technique used to observe events that are located in or close to the plasma membrane of a cell. By using an evanescent field for fluorochrome excitation which only penetrates 60-250 nm into the cell, TIRF microscopy provides an unmatched z-resolution which enables the imaging of events occurring in or close to the plasma membrane, e.g., molecule transport to the plasma membrane. TIRF avoids the problem of being overwhelmed by fluorescence signals from molecules deeper inside the cell.
TIRF images: Transport of Galectin-3 vesicles close to the membrane along actin filaments.
YFP: red; CFP: green; scale of overview images: 20 µm; section thickness: 6 µm; penetration depth TIRF: 110 nm. Courtesy of Ralf Jacob, University of Marburg, Germany.
FRET and BRET– quantifying protein-protein interactions
Förster resonance energy transfer or FRET belongs to a group of photomanipulation techniques that enable you to interact with the observed event in the specimen through the use of light.
FRET is a useful tool for quantifying molecular dynamics, such as protein-protein interactions, protein-DNA interactions, and protein conformational changes. FRET imaging normally uses derivatives of GFP (green fluorescent proteins), particularly CFP and YFP (cyan and yellow fluorescent protein, respectively), which are each attached to proteins of interest using molecular biology methods. Then the CFP molecule is excited with fluorescent light. As soon as the proteins of interest are in close spatial proximity (<20 nm), the CFP will act as a donor and transfer the energy which is emitted in the form of light to the YFP which acts as an acceptor. The researcher will observe a shift from blue fluorescence emitted from the CFP to yellow fluorescence emitted from the YFP. In the case of BRET (bioluminescence resonance energy transfer) the donor is a bioluminescent molecule (e.g. luciferase derivatives) that acts as the donor and, like in FRET, a GFP derivative acts as the acceptor.
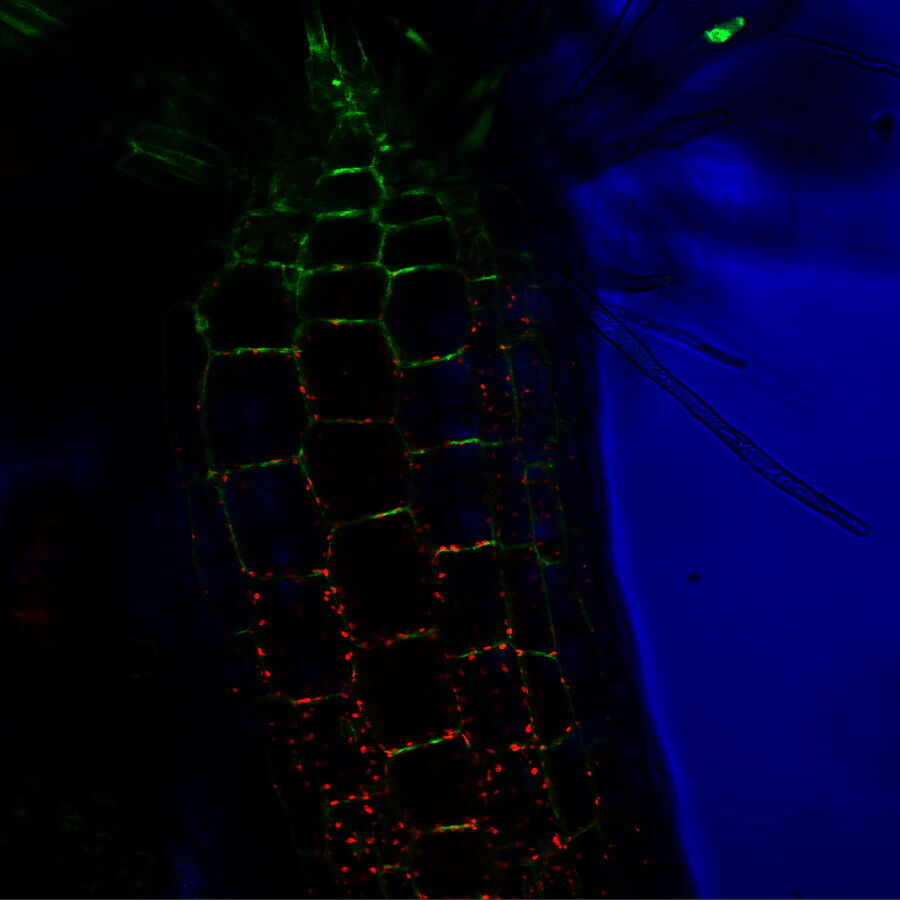
FRAP – monitoring protein and vesicle trafficking
Fluorescence recovery after photo-bleaching (FRAP) is another photomanipulation technique often used to monitor protein or vesicle trafficking. FRAP uses a fluorescent protein (usually GFP) attached to a protein of interest, i.e., a protein whose movement is to be monitored. Usually, the whole cell is initially fluorescent as the protein might be abundant throughout the whole cell. Then a certain region of the cell, often with cell structures like axons or dendrites in neuronal cells, is exposed to high intensities of light to eliminate (bleach) the fluorescence in that particular region. As the protein of interest moves, it will appear in the bleached region at a certain speed, which can be traced with the recovering fluorescent signal from this region, giving researchers insight into intracellular transport dynamics.
Photoactivation – monitoring gene expression and protein transport
Photoactivation, a recently developed method, selectively labels areas of interest within a cell or organism by using especially designed dyes, such as photoactivated-green-fluorescent protein (paGFP) or Kaede. These dyes only show significant fluorescence after illumination with light of a specific wavelength. These dyes can also be fused to a protein of interest to study its expression or transport dynamics by using FRAP or particle tracking.
Conclusions
Live-cell imaging is an indispensable tool in life science research that allows the visualization of cells in a state that is as close to in vivo as possible. Acquisition of images of living cells enables you to fully understand and address your research questions regarding cell movement, growth, and dynamic processes.
The future is quantitative
Biological research has moved from purely descriptive investigation and entered an era of quantitative analysis. New live-cell imaging techniques are evolving in the direction of higher resolution, both in space and time. Technological developments are now concentrating on the quantitative study of dynamic events, making these methods more accessible and fundamental to fully answer research questions.