Hyperplex Cancer Tissue Analysis at Single Cell Level with Cell DIVE
How multiplexed imaging of single cells provides insights into tumor microenvironments

The ability to study how lymphoma cell heterogeneity is influenced by the cells’ response to their microenvironment, especially at the mutational, transcriptomic, and protein levels. Protein expression studies offer the most relevant information about the nature of cellular interactions and protein expression levels. A hyperplexed workflow can be applied for studying multiple proteins from the same cancer tissue.
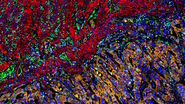
Image: Colon adenocarcinoma. 13 biomarkers shown: Cadherin, CD3, CD4, CD8, CD20, CD31, CD45, Collagen, Caspase 9, BCL2, Beta-catenin, Vimentin and SMA.
What to expect in the webinar
With Cell DIVE™ technology, researchers can investigate crucial topics such as lymphoma cell heterogeneity and the nature of relapses and therapeutic resistance—in the context of immunological synapse spatial organization.
Key Learnings
- How the hyperplex workflow can be applied for studying multiple proteins from the same cancer tissue
- More about the technical aspects of the hyperplex workflow
- How Cell DIVE technology enables spatial profiling
- Why image analysis in immunooncology is important