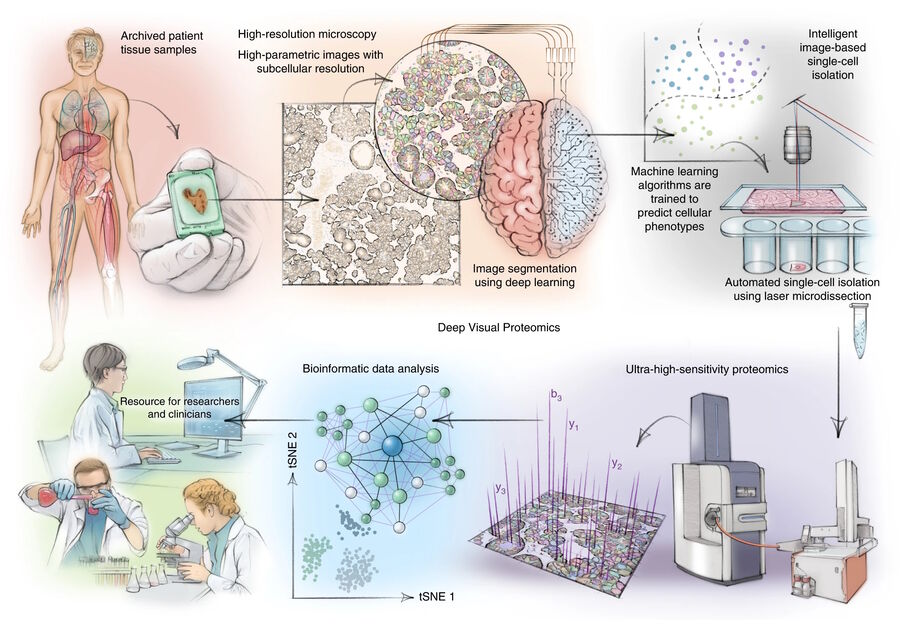
About Deep Visual Proteomics (DVP)
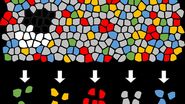
DVP [1] links high-resolution, data-rich imaging of cell cultures or biobank tissues with deep-learning-based cell segmentation and machine-learning-based identification of cell types and states. Cellular or subcellular features of interest classified by artificial intelligence (AI) undergo automated laser microdissection (LMD) [1] and proteomic profiling via mass spectroscopy (MS) [2]. Subsequent bioinformatics data analysis enables data mining and the discovery of protein signatures, providing molecular insights into proteome variation concerning health and disease states at the single-cell level. The DVP concept and workflow is shown in figure 1.
Identifying single cells for cellular heterogeneity
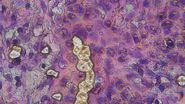
By individually excising nuclei from cell culture, distinct cell states can be classified with proteomic profiles defined by known and uncharacterized proteins. In an archived primary melanoma tissue, DVP identified spatially resolved proteome changes as normal melanocytes transition to fully invasive melanoma. It revealed pathways that change in a spatial manner as cancer progresses, such as mRNA splicing dysregulation in metastatic vertical growth that coincides with reduced interferon signaling and antigen presentation. The ability of DVP to retain precise spatial proteomic information in the tissue context has implications for the molecular profiling of clinical samples.
For this research, cells or nuclei were excised using a LMD7 laser microdissection microscope which was adapted for automated single-cell automation [1] (refer to figures 2-4).
Utilizing DVP with the Leica LMD system to discover therapeutic targets for fatal drug reactions
Two years after its introduction in Nature Biotechnology, Deep Visual Proteomics (DVP) has achieved a major breakthrough by identifying a therapeutic target for Toxic Epidermal Necrolysis (TEN), a severe skin reaction triggered by common medications. TEN patients experience life-threatening epidermal detachment, creating an urgent need for targeted therapies. In a recent study, Nordmann et al. utilized the DVP workflow, powered by the Leica LMD system for precise laser microdissection, alongside AI-driven cell segmentation and ultra-sensitive mass spectrometry, to dissect and analyze skin biopsies from TEN patients at the cellular level.
The Leica LMD system enabled accurate isolation of single keratinocytes and immune cells, allowing the quantification of over 5,000 proteins. This analysis revealed elevated type I and II interferon signatures and activated phosphorylated STAT1. Further exploration of the JAK/STAT and interferon pathways suggested that JAK inhibition (JAKi) could mitigate TEN severity. Remarkably, JAKi treatments led to significant improvements, as seen in rapid re-epithelialization in seven TEN patients following initial success in mouse models. This discovery highlights the power of the DVP workflow, with the Leica LMD system at its core, in identifying critical therapeutic options and offering hope for effective treatment strategies for TEN and other severe immune-mediated conditions.
To find out more about using DVP, with AI-guided imaging, LMD, and MS, for identifying single cells, read the full article:
A. Mund, F. Coscia, A. Kriston, R. Hollandi, F. Kovács, A.-D. Brunner, E. Migh, L. Schweizer, A. Santos, M. Bzorek, S. Naimy, L.M. Rahbek-Gjerdrum, B. Dyring-Andersen, J. Bulkescher, C. Lukas, M.A. Eckert, E. Lengyel, C. Gnann, E. Lundberg, P. Horvath, M. Mann:
Deep Visual Proteomics defines single-cell identity and heterogeneity
Nature Biotechnology (2022) vol. 40, pp.1231–1240
DOI: 10.1038/s41587-022-01302-5
https://www.nature.com/articles/s41587-022-01302-5
---
Read how DVP was utilized to find cure against a fatal drug reaction:
TM. Nordmann, H. Anderton, A. Hasegawa, L. Schweizer, P. Zhang, P. Stadler, A. Sinha, A. Metousis, FA. Rosenberger, M. Zwiebel, TK. Satoh, F. Anzengruber, MT. Strauss, MC. Tanzer, Y. Saito, T. Gong, M. Thielert, H. Kimura, N. Silke, EH. Rodriguez, G. Restivo, HH. Nguyen, A. Gross, L. Feldmeyer, L. Joerg, MP. Levesque, PJ. Murray, S. Ingen-Housz-Oro, A. Mund, R. Abe, J. Silke, C. Ji, LE. French & M. Mann
Spatial proteomics identifies JAKi as treatment for a lethal skin disease
Nature (2024)
https://www.nature.com/articles/s41586-024-08061-0