Achieving in-depth insights into the tumor microenvironment
Understanding the nature of cancer genesis and progression requires understanding the fine details of the tumor microenvironment, or TME. However, due to the complexity of the TME, with its many tissues, cell types, and dynamics many traditional cell biology tools fall short of capturing the complete map of the terrain. Spatial biology approaches are crucial for understanding the intricate spatial relationships between these cells, tissues, and molecular components, which in turn provides valuable insights into disease progression, treatment response, and the development of targeted therapeutic interventions. by studying multiple biomarker channels and visualizing them in a single sample using multiplex spatial imaging, we can gain a comprehensive understanding of the complex interplay between different biomarkers within a biological system. Traditional widefield microscopy can be limited in the number of biomarkers that can be simultaneously imaged. Due to the spectral overlap and limitations in available fluorophore combinations, it becomes challenging to visualize and distinguish a large number of biomarkers (i.e. more than 6-7) within the same sample. This poses a significant constraint when studying highly complex biological systems that involve numerous biomarkers and intricate molecular interactions.
Cell DIVE-IBEX hybrid method: Advancing multiplex imaging in research
One end-to-end solution that helps users perform this type of imaging is Cell DIVE. Cell DIVE uses a patented, gentle, tissue-preserving dye inactivation solution to allow for the iterative staining of 60+ biomarkers in a sample. However, there are alternative chemistries and methods to accomplish similar imaging techniques. One such popular method in the scientific literature is known as IBEX (Iterative Bleaching Extends Multiplexity) imaging. It was invented by the laboratory of Ronald Germain at the National Institutes of Health in Bethesda, Maryland. IBEX is designed as a non-commercial solution for iterative multiplexing and offers a list of a wide variety of compatible antibodies (250+), dyes (16+), as well as software packages to streamline image acquisition. However, IBEX users may be interested in leveraging the workflow simplification, process automation, software, and image processing tools that Cell DIVE has to offer.
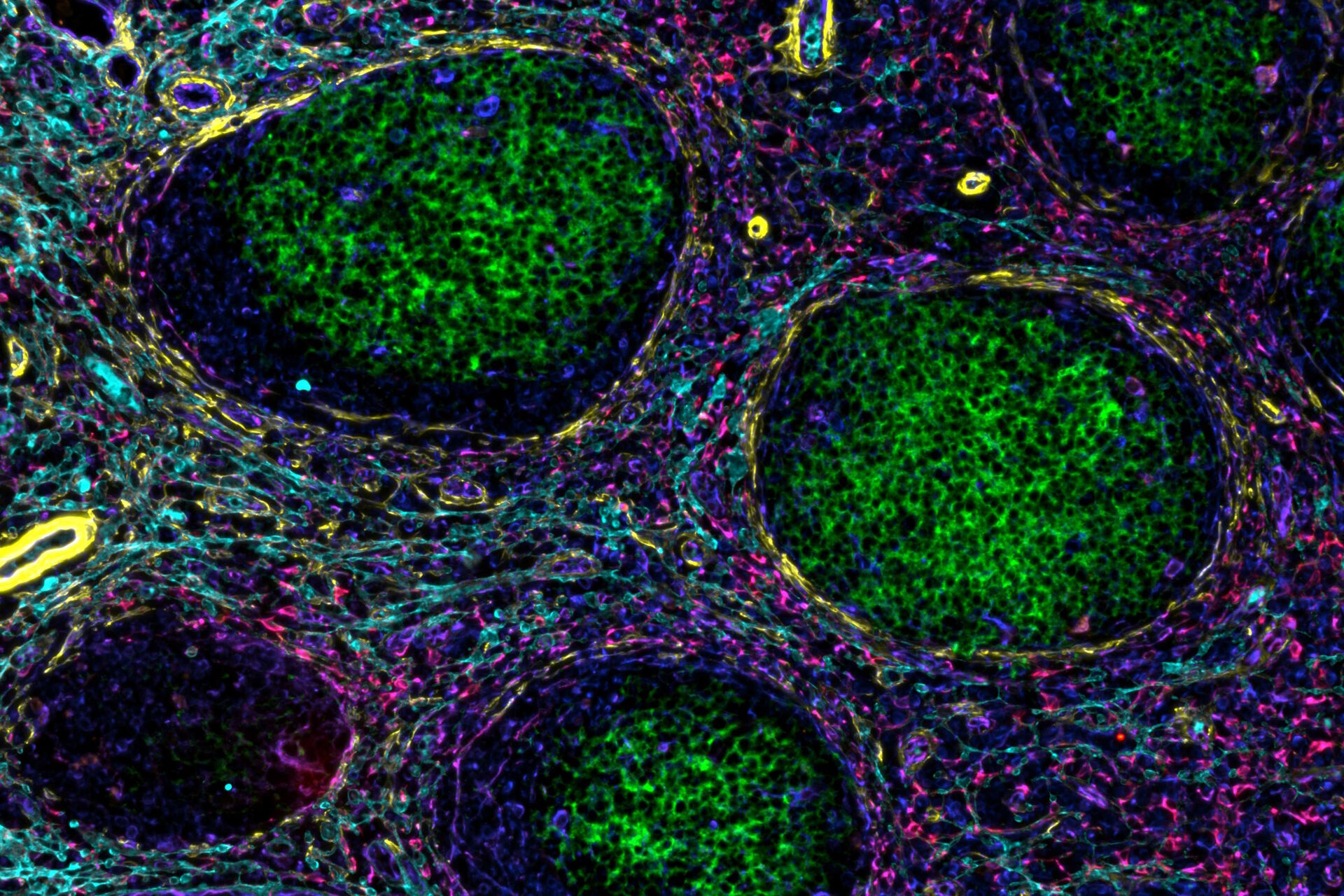
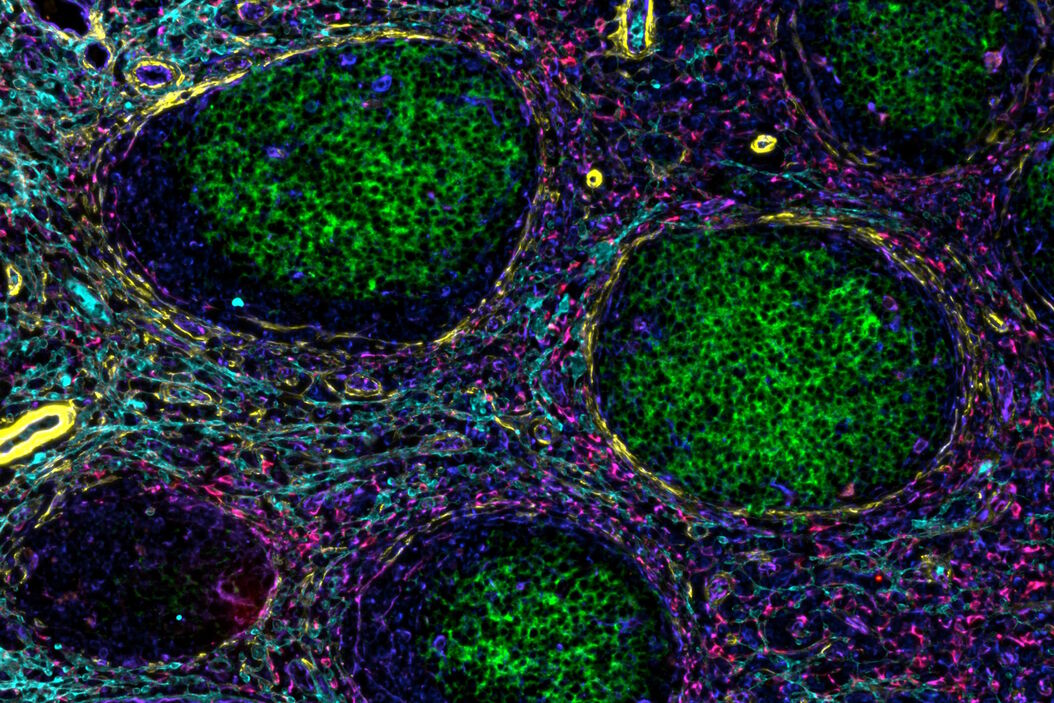
In a recent publication by Andrea Radtke and others from the Germain laboratory, a novel, hybrid method entitled Cell DIVE-IBEX establishes that such a notion is readily achievable. In broad terms, antigen retrieval, antibody staining, and dye inactivation methods pioneered for IBEX were used to process samples to be imaged utilizing the Cell DIVE imager and software. This allowed the scale of part of the study to be increased over 4-fold to dig deeper into the questions surrounding the progression of follicular lymphoma in patient samples.