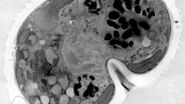
Case study: using organoids as a model system to study cancer development
Edoardo D'Imprima from the EMBL presents his research on imaging cancer development in organoids, which bridges the gap between 2D cell cultures and animal models. Organoids offer scalability for studying cancer development across nanometer to millimeter scale. The challenge lies in integrating live cell imaging microscopy with cryo-electron microscopy to visualize molecular complexes at atomic resolution. Combining the strengths of both techniques, light microscopy provides fluorescence information about molecules of interest (e.g., localization) while electron microscopy offers higher resolution. Volume electron microscopy allows precise imaging of organoid structures, aiding in the understanding of cancer development. This approach has been demonstrated successfully on patient-derived organoids, showcasing detailed imaging of cellular structures like mitochondria and centrioles.
The webinar was broadcast in September 2023.