Virus-membrane interactions
A well established usage case for cryo-TEM is three-dimensional reconstruction of isolated macromolecules, virus particles, or filaments. On one hand, these approaches are based on averaging of repetitive structures – either due to numerous identical molecules, repetitive patterns on a filament, or symmetries, to reduce the noise inherent to cryo-TEM. On the other hand, different views of these complexes required for three-dimensional reconstruction can be either obtained from (ideally) random orientation of the particles in the ice or from the helicality of filaments.
Mohit Kumar from Dieter Blaas' Group at the Max F. Perutz Laboratories (Vienna) was interested in the interaction of rhinovirus particles with lipid membranes and the events leading to endocytosis of the particles [1]. They decided to employ single particle reconstruction methods to obtain three-dimensional models of the particles docking to artificial membranes (liposomes), using numerous individual virion/membrane complexes selected from many micrographs.
For collection of many micrographs, as required by this type of experiment, or for freezing of numerous grids of similar specimens, reproducibility of results – in this case ice thickness and minimized contamination – is a major factor. The Leica EM GP addresses problems with blotting intensity arising from moist, flexing filter paper or bent grids with a unique sensor: this sensor detects the moment of contact between the filter paper and the sample droplet, establishing a reference point. By moving the filter paper forward by a defined distance from this reference point, the pressure applied varies very little from one grid to the next, yielding highly reproducible results if the same specimen, volume and timing are being used.
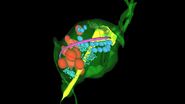
Using this blotting sensor, results could be obtained very efficiently from 4 µl samples applied onto glow discharged 400 mesh Cu grids with a R1.2/1.3 Quantifoil film that were allowed to spread for 25 seconds in the environmental chamber with 99 % humidity and 30 °C. Subsequently, they were blotted for 0.8 seconds and plunged into liquid ethane. Particles binding to liposomes of similar diameter were picked from the micrographs of these frozen and unstained specimens acquired on a 300 kV cryo-TEM (Figure 1). From these selected particles, the authors calculated reconstructions of the virus/membrane assemblies in absence of cellular receptors, mimicking the intimate contact of lipid membranes over a twofold axis of icosahedral symmetry, illustrating the binding mechanism of the tight virus-membrane complex (Figure 2).
Fig. 2: Three dimensional reconstruction, computed via averaging 275 single particle images extracted from micrographs as exemplified in Fig. 1. Binding to the membrane is seen to occur close to one of the 2-fold axes of icosahedral symmetry. The lipid bilayer was localized via asymmetric 3DR yielding a poorly resolved virion; to aid identification of its axes, the volume was symmetrized and superimposed (courtesy of Mohit Kumar and Dieter Blaas, MFPL, Vienna/Austria).
Baculovirus actin comet tails
Jan Mueller and colleagues from J. Victor Small's lab at the Institute of Molecular Biotechnology, Vienna [2] used cryo-electron tomography to investigate the actin cytoskeleton in baculovirus "comet tails", both in vivo as well as in vitro. These actin tails are nucleated by baculovirus and other pathogens to drive their own propulsion, essential to disseminate the infection. They serve as a popular and well accessible model systems to study the architecture of the actin cytoskeleton and the role of regulatory proteins.
For the in vitro experiments with actin tails polymerized from synthetic (and easy to manipulate) "motility cocktails", the tails were not pipetted onto the grids just before freezing, to prevent damage by manipulation. Instead, they were "grown" directly on Quantifoil R3.5/1 perforated carbon films on 200 mesh Au grids. After supplementing the sample with 10 nm protein coated colloidal gold serving as fiducial markers for alignment in electron tomography, it was transferred to the humidified chamber of the EM GP. Samples were blotted 1.2 to 1.6 s from the back side, subsequently frozen by plunging into liquid ethane just above freezing temperature, and stored under liquid nitrogen until loaded into the microscope. To reduce background, zero-loss filtered tilt series were acquired with SerialEM over the holes of the perforated carbon film at 300 kV and reconstructed offline with IMOD (Figure 3).
For another part of the study, baculovirus actin comet tails were visualized by Mueller and colleagues inside the thinly spread part of living cells that is is accessible to examination by cryo-electron microscopy. The investigation of this region was greatly advanced by the introduction of cryo-electron tomography and the ability to resolve superimposing structures in three dimensions [3].
For these in vivo experiments, cells were cultured on 200 mesh gold grids, to eliminate cytotoxic effects by copper ions and obstruction by grid bars at high tilts in electron tomography. A perforated carbon support film such as Quantifoil R1/4 was used. Once the cells had attached and spread, they were chemically fixed and extracted to clear the cytosolic background, enabling reliable tracking of individual filaments. Subsequently, samples were covered with 4 µl of medium supplemented with 10 nm protein saturated gold particles and transferred to the environmental chamber of the EM GP at 95 % relative humidity. These samples benefited greatly from the design of the blotting mechanism on the Leica EM GP: due to unilateral blotting, excess liquid was only blotted away from the back side of the grid, direct contact between the sensitive cell monolayer and the filter paper is avoided. Blotting times of 1.5 to 2.0 s with pre-humidified Whatman No. 1 filter paper yielded best results, using the holes in the C film to obtain an even ice thickness all over the grid. After blotting, the samples were plunged immediately into liquid ethane. Tomography data were acquired and processed as described above (Figure 4).
Fig. 3: Cryo-electron tomography of baculovirus actin comet tail in vitro. a) Cryo-electron tomogram section (11 nm) of a comet tail formed on a purified, de-enveloped baculovirus in vitro in a motility cocktail containing actin, Arp2/3 complex, gelsolin, cofilin, and VASP. Insets show details of branch junctions from the squares in the overview image. b) Model derived from tomogram, showing actin filaments in green and branch junctions in red. c) Close-up of the model highlighting the filaments abutting the rear of the virus in yellow. Bars (a, b), 100 nm; c) 25 nm; inset, 10 nm (Mueller J et al.: Electron Tomography and Simulation of Baculovirus Actin Comet Tails Support a Tethered Filament Model of Pathogen Propulsion. PLoS Biol. 12 [1]: e1001765, Figures a–c; doi: 10.1371/journal.pbio.1001765.g005).
Fig. 4: Cryo-electron tomography of a baculovirus actin comet tail in vivo. a) Cryo-electron tomogram of a baculovirus comet tail in a B16 melanoma cell. Image shows 19 nm sections of the tomogram. Since the virus tail was not in one plane in the ice layer, the tomogram is shown in three images, separated by white lines, taken at different z-levels. Insets show details of branch junctions from the squares in the overview image. b) Projection from the rear of the complete comet tail model with actin filaments of the host cytoskeleton (translucent) as well as one microtubule (grey tube) superimposed. Bar a), 100 nm (Mueller J et al.: Electron Tomography and Simulation of Baculovirus Actin Comet Tails Support a Tethered Filament Model of Pathogen Propulsion. PLoS Biol. 12 [1]: Figures 3a + d; doi: 10.1371/journal.pbio.1001765.g003).
Pharmaceutical nanocarriers
Immersion freezing and cryo-TEM are not only indispensable tools in structural and cellular biology, but also very useful for imaging of pharmacological specimens in their natural state (for a review, see [4]). A study by [5] used cryo-TEM of immersion frozen specimens to elucidate the ultrastructure of different ultrasound-engineered nanocarriers for dermal drug delivery: solid lipid nanoparticles, nano-structured lipid carriers, and nanoemulsions were visualised.
Samples for these experiments were frozen on a Leica EM GP, the instrument’s environmental chamber was operated at room temperature and 95 % rH. The optimal dilution of each specimen in double distilled water was obtained from a series of pre-tests, to yield a good density of specimen with not too much overlap between individual structures. 4 µl of the specimen were applied onto a glow discharged 400 mesh EM grid coated with perforated carbon films. After a settling time, the suspension was blotted for 1.25–4.0 s, depending on the formulation used. Standard Whatman No. 1 filter paper and the instrument’s blotting sensor were used, the sample was immediately plunged into liquid ethane after blotting.
While size information from cryo-TEM was complementary to data obtained form dynamic light scattering, structural information was exclusive to cryo-TEM and showed quite dramatic differences between individual specimens, ranging from spherical droplets to flat, flake-like structures (Figure 5). Please note the absence of contamination in the micrographs presented here, owing to quick handling and a number of anti-contamination measures integrated in the GP: this includes negative pressure in the environmental and a powerful "TF" evaporator in the primary cryogen, that creates an atmosphere of dry and cold N2 gas, to protect the specimens from warming up and contamination.
Fig. 5: Cryo electron microscopic images of nano-structural lipid carriers (NLC), nanoemulsions (NE) and solid lipid nanoparticles (SLN) (Schwarz JC et al.: Nanocarriers for dermal drug delivery: Influence of preparation method, carrier type and rheological properties. International Journal of Pharmaceutics 437: 83–88 [2012], Fig. 2).
Recommended reading
- Resch GP, Brandstetter M, Königsmaier L, Urban E, and Pickl-Herk AM: Immersion freezing of suspended particles and cells for cryo-electron microscopy. Cold Spring Harb. Protoc. 803–14 (2011).
- Resch GP, Brandstetter M, Wonesch VI, and Urban E: Immersion freezing of cell monolayers for cryo-electron tomography. Cold Spring Harb. Protoc. 815–23 (2011).
References
- Kumar M, and Blaas D: Human rhinovirus subviral a particle binds to lipid membranes over a twofold axis of icosahedral symmetry. J. Virol. 87: 11309–12 (2013).
- Mueller J et al.: Electron tomography and simulation of baculovirus actin comet tails support a tethered filament model of pathogen propulsion. PLoS Biol. 12: e1001765 (2014).
- Medalia O et al.: Macromolecular architecture in eukaryotic cells visualized by cryoelectron tomography. Science 298: 1209–13 (2002).
- Klang V, Valenta C, and Matsko NB: Electron microscopy of pharmaceutical systems. Micron 44: 45–74 (2013).
- Schwarz JC et al.: Nanocarriers for dermal drug delivery: influence of preparation method, carrier type and rheological properties. Int. J. Pharm. 437: 83–88 (2012).