Laser microdissection and molecular biological methods
Particularly for molecular biological analysis methods, such as quantitative PCR, successful examination depends on maximum precision and freedom from contamination. The contact-free isolation and separation offered by the laser microdissection method is especially suitable for extracting the following structures:
- Single cells from tissue samples;
- Cell components;
- Areas of tissue;
- Chromosomes; and
- Live cells from cell cultures.
Extraction of pure material
Once successfully removed by the laser microdissection technique, the dissectates can be subjected to molecular biological and biochemical methods, such as nucleic acid analysis and protein investigations. In particular, LMD dissectates can be used with established analysis techniques, like PCR (polymerase chain reaction), real-time PCR (quantitative PCR), Southern and Western blots, cloning, RNA analysis in the form of total and specific mRNA isolation, reverse transcription PCR, Northern blots and also protein analysis in the form of 2-D SDS-PAGE (sodium dodecyl sulfate polyacrylamide gel electrophoresis), LC-MS (liquid chromatography mass spectrometry).
The following describes the use of laser microdissection in molecular biology – embracing the fields of DNA and RNA analysis and proteomics – in more detail. Apart from these highlighted applications, laser microdissection is also an effective technique for cell culture, plant research, forensics, and climate research.
DNA research
Laser microdissection is the method of choice for extracting even extremely small samples from specimens for molecular biological investigation. After dissecting the target area by laser, it is possible to isolate and then duplicate the nucleic acid it contains. A special variant of PCR called "lab on a chip" is extremely useful for this purpose.
DNA analysis is performed in molecular biology to clarify such things as medical and diagnostic issues. It requires the duplication of genetic material by PCR variants after cell lysis. PCR is based on the principle of specific amplification of distinct sequence sections of DNA strands. One of the uses of laser microdissection with subsequent DNA amplification is the detection of diseases and genetic modifications.
Another application that has rapidly gained significance in the most diverse areas of clinical medicine and molecular biological research is the examination of single cells. Many questions can only be answered by specific analysis of single or multiple cells and the use of a range of molecular biological methods. Laser microdissection serves a specific purpose for the extraction and examination of single cells.
One of the greatest problems in oncology is the selective isolation of DNA from cancer cells in growing tumors. In the early stage, the affected area of tissue is often only small and little material is available. One type of solution is to pre-select the relevant cells using the laser microdissection technique, then isolate and compare them with healthy surrounding tissue to look for signs of mutation.
RNA research
An examination of genetic information using molecular biological and biochemical methods is termed gene-expression analysis. It provides both quantitative and qualitative information about genetic activity. Microarray technology has become increasingly widespread in genome and gene expression analysis and diagnostics. By means of these so-called gene chips, it is possible to examine the expression of many genes at the same time.
Laser microdissection is used to generate a cell-specific gene expression profile of a single or multiple cells of the same type. Laser microdissection plays a special role in neurosciences for removing individual neurons from tissue, in particular to separate them from the surrounding glia cells. The technique is equally successful for precisely cutting through axons or dissecting synapses.
For dopamine research, the focus is on gene expression analysis of dopamine-producing neurons in the midbrain. Malfunctions of the so-called dopamine midbrain system are observed in disease patterns, such as Parkinson’s disease, schizophrenia, ADHD (attention-deficit / hyperactivity disorder), and drug addiction. Isolating single cells in both healthy and diseased areas of tissue is necessary when it comes to progressive, selective cell losses, as is the case for Parkinson’s disease.
Laser microdissection enables individually affected cells to be removed from the substantia nigra of Parkinson’s patients and from non-affected, healthy control tissue in order to detect differences in the gene expression of these cells during the development of the disease. The gene expression of dopaminergic substantia nigra neurons can also be compared with dopaminergic neurons from the neighboring ventral tegmental area, which is less vulnerable to Parkinson’s, in order to find out what makes the neurons more vulnerable or resistant.
Proteomics
Proteomics is concerned with structure elucidation and quantitative analysis of protein expression and interactions. It provides information on components of metabolic pathways and regulatory circuits. Thus, it supplements and validates the data obtained from gene expression analysis.
Laser microdissection is useful for selecting and isolating cells in preparation for proteomic analysis. Techniques, such as MALDI-TOF IMS (matrix-assisted laser desorption/ionization time-of-flight imaging mass spectrometry) or MALDI-FTICR (Fourier transform ion cyclotron resonance) IMS, have been applied for drug development. A longitudinal section of a single human hair obtained through laser microdissection can be directly tested for methamphetamine with the MALDI-TOF-IMS technique.
Deep visual proteomics
One subcategory of proteomics is spatial proteomics, where protein abundance is correlated with cellular or subcellular locations. Especially, artificial-intelligence-driven image analysis of cellular phenotypes with automated single-cell or single-nucleus laser microdissection and ultra-high-sensitive mass spectrometry, which is called Deep Visual Proteomics (DVP), should be mentioned here [1].
Cell cultures
The special advantage of using laser microdissection for live cells is the possibility to damage specific cells with the laser in order to be able to observe their subsequent regeneration. It is also possible, depending on the focus of research, to use laser microdissection to isolate selected areas or distinct clones from live cell cultures for further cultivation or additional analysis, such as PCR.
This approach means that the relevant region can be examined without the surroundings falsifying the result. Such techniques are also suitable for cell surgery and similar manipulations (see below). Even sensitive stem cells can be selected with laser microdissection without losing their division potential. Suitable for a wide range of research purposes, these cells can be used for future applications in stem-cell therapy, regenerative medicine, and drug screening.
Plant research
Research of plant tissue on a molecular level poses an enormous challenge. The main obstacle to overcome in sample preparation is the woody stems and thick cell walls of plant tissue compared to animal tissue. The laser used in the Leica LMD system is able to cut through even thick and hard plant material. By obtaining information at different cellular levels, it is possible to gain a better understanding of plant reactions.
Relevant issues in this context are development and differentiation processes of plant tissue, their metabolism, adaption, development of diseases, and resilience. Other interesting aspects are symbioses and reactions of plants to bacteria, viruses, fungi, and parasites.
Laser microdissection helps to produce a clearer picture of plant cells. The information gained could contribute towards an increase in biomass production, for instance, or plant-disease control. Laser microdissection can also be used to isolate colored leaf pigments for subsequent NMR-spectrometric analysis.
Forensics
One part of forensics is genetic fingerprinting which plays a key role in many areas today, particularly in crime investigation. A particular challenge faced by forensic scientists are impurities of all kinds in samples. Using laser microdissection, it is now possible to separate the required cells from impurities. It is also possible to distinguish between male and female cells applying new cell-identification processes and fluorescence illumination. These cells can be quickly identified and isolated through laser microdissection.
Laser microdissection plays a particularly important role for isolating sperm from smear preparations. Single sperm cells are identified, separated from skin cells and impurities, and then analyzed.
Additional Applications
Climate research
One additional application where laser microdissection can be used is climate research. Thanks to the powerful laser of the Leica LMD system, it is possible to section through relatively hard materials, such as wood. It can be used to isolate annual rings or inclusions from thin sections of wood.
CLEM and LMD
The increasing complexity of protein and organelle investigations in cell biology sometimes demands a combination of different imaging approaches. The CLEM method in particular couples together technologies like light and electron microscopy. The key aim is to display optical microscope images at the level of electron microscopy, i.e., to correlate dynamic events in living cells with ultrastructural and 3D information. Using LMD, a system of reference coordinates can be produced on the surface of a culture substrate to enable the location of labeled details to be determined quickly and precisely during sample preparation. This process is impressively illustrated in Figure 5.

NanoSIMS
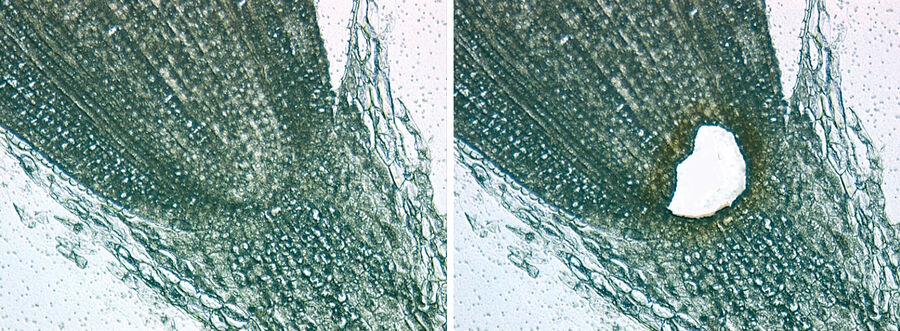
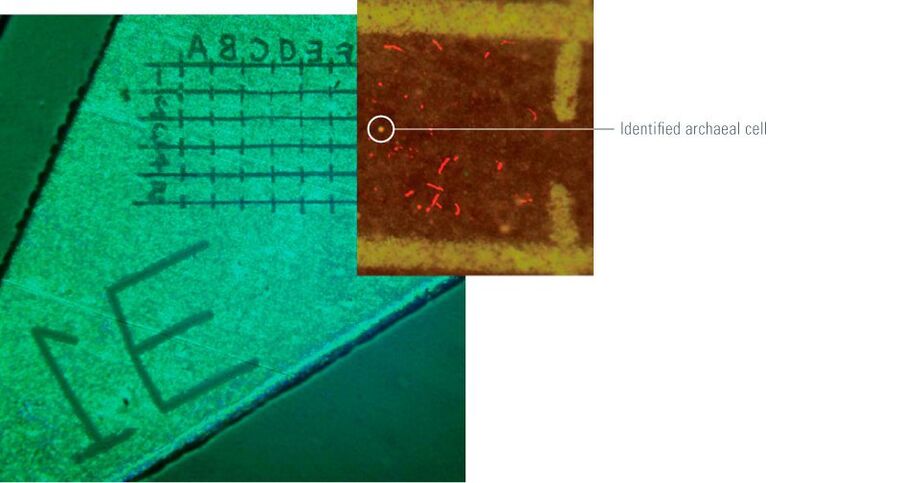
Bacteria analysis is performed using NanoSIMS (nanometer-scale secondary ion mass spectrometry) coupled with LMD and AFM (atomic force microscopy) to obtain a three-dimensional image showing the element and isotopic compositions of a sample with a spatial resolution of <50 nm. The LMD method proves to be advantageous here, as the laser produces a grid pattern on the surface of the filters in use to mark structures of interest (Figure 6). This method is used in marine biology, for example, to analyze carbon and nitrogen fixation in the Pacific [3, 4] and Baltic Sea [5].
Live-Cell Manipulation
The Leica LMD system is also especially useful for the live-cell sector in terms of the deliberate mechanical destruction of cellular structures, such as centrosomes, microtubules, and membranes. For instance, cell membranes can be perforated with the laser to allow permeation of substances that were previously completely blocked by the membrane, according to Prof. Monica Gotta from the Department of Genetic Medicine and Development at the University of Geneva, Switzerland. Most importantly, it is possible to manipulate the spindle apparatus of cells during cell division (Figure 7).
![Mitotic spindle ablation in a wild type one-cell embryo. Fig. 7: Mitotic spindle ablation in a wild type one-cell embryo. A) C. elegans one-cell embryo expressing α-tubulin fused to the GFP. The Mitotic spindle just before (a) and after laser ablation (b). The two spindle poles separate after the ablation [6].](/fileadmin/_migrated/pics/Mitotic_spindle_ablation_in_a_wild_type_one-cell_embryo.jpg)
Current publications using laser microdissection
Links to current papers discussing the most common applications for laser microdissection can be found in this publication list:
https://www.leica-microsystems.com/products/light-microscopes/p/leica-lmd7/